Pattern matching language and tools
Developed at IRISA, a French academic research institute, Logol is a pattern matching grammar language and a set of tools to search a pattern in a sequence (nucleic or proteic).
It includes a Linux command line tool and a graphical designer.
Software is free and open source, under CeCILL license
You can also use it on the GenOuest platform online (with restrictions) or via command-line with a GenOuest account.
Download
Sample code
mod1()==*>SEQ1
mod1()==>"aaa","acgt":{_R1},?R1
Download a full example (grammar+result file)
Sample explanation
This grammar searches in a sequence the term "aaa", then the term "acgt" saved as variable R1, and at least the content of variable R1 ("acgt" again).Features
- Exact, approximate, local and remote search
- Hamming and distance errors support
- Unlimited variables use
- Transformations (reverse, complement, custom)
- Contraints: position, size, length,...
- Alphabet percentage
- And much more!
Training
We provide an extensive documentation to learn Logol, but we can also provide on-site training.
You can contact us at
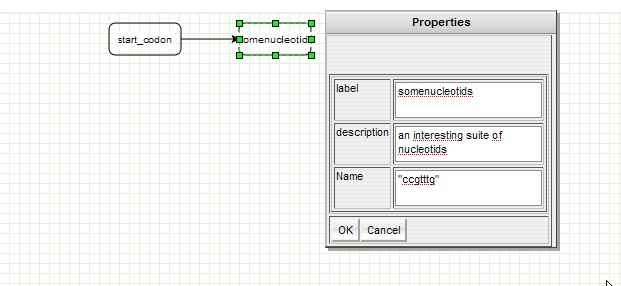
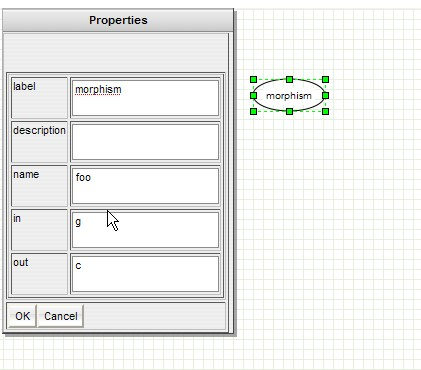
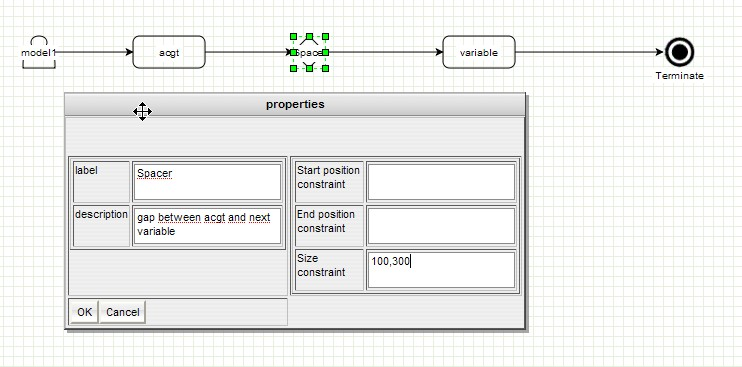
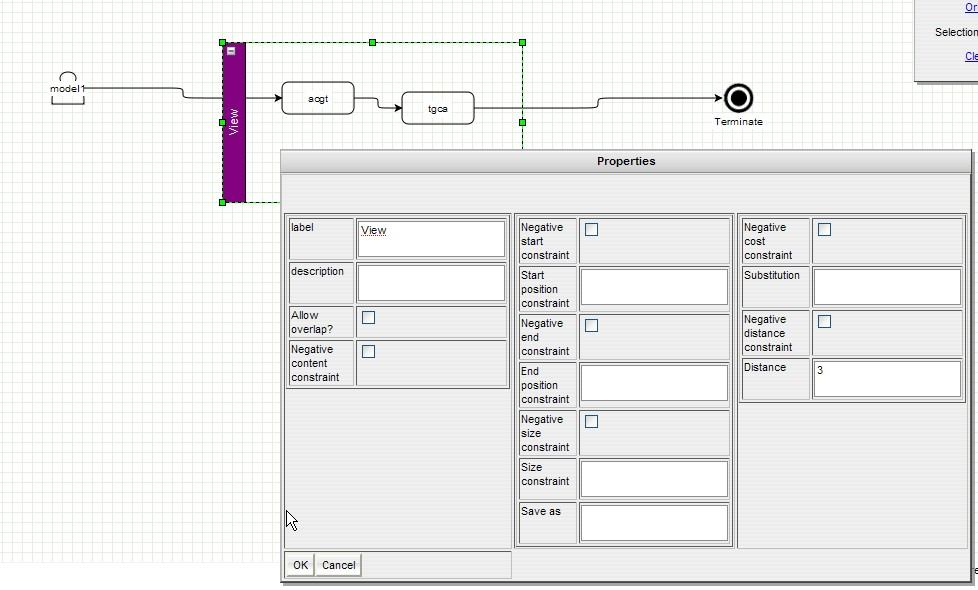
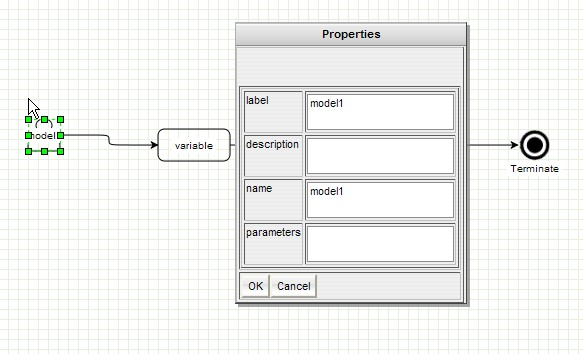
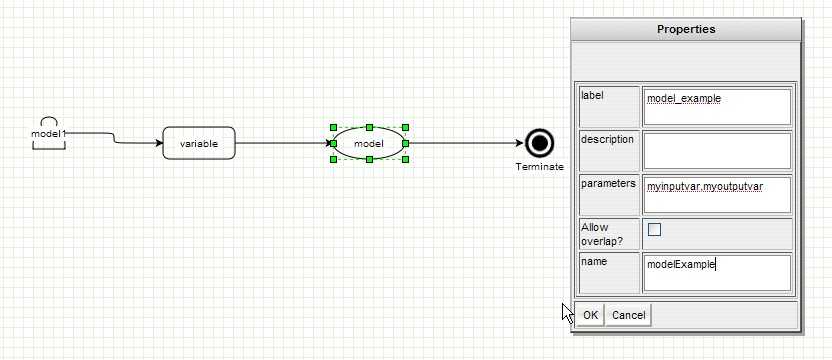
GenOuest support:support@genouest.org.Graphical editor screenshots
Authors:
Olivier Sallou (Univ. Rennes 1)
Catherine Belleannée (Univ. Rennes 1)
Jacques Nicolas (INRIA)
Olivier Sallou (Univ. Rennes 1)
Catherine Belleannée (Univ. Rennes 1)
Jacques Nicolas (INRIA)
Contact:
GenOuest Platform, IRISA
Campus de Beaulieu
35000 Rennes
France
GenOuest Platform, IRISA
Campus de Beaulieu
35000 Rennes
France
Credits:
Theme by Twitter bootstrap
Fonts by http://www.fontspace.com/rasdesign/rayando
Icons by Glyphicons
Theme by Twitter bootstrap
Fonts by http://www.fontspace.com/rasdesign/rayando
Icons by Glyphicons